Quantum-Classical Hybrid Neural Network Comparison
This notebook compares different neural network architectures including:
Classical Multi-Layer Perceptrons (MLP)
Quantum circuits with LINEAR output mapping
Quantum circuits with various GROUPING strategies
1. Import Required Libraries
First, we’ll import all necessary libraries for our experiment:
PyTorch: For neural network implementation and training
Perceval: For quantum circuit simulation
Matplotlib/Seaborn: For visualization
Custom modules: For quantum layers and MLP models
[ ]:
from collections import defaultdict
import matplotlib.pyplot as plt
import perceval as pcvl
import seaborn as sns
import torch
import torch.nn as nn
from sklearn.metrics import accuracy_score, classification_report, confusion_matrix
from tqdm import tqdm
%matplotlib inline
from merlin import (
ComputationSpace,
LexGrouping,
MeasurementStrategy,
ModGrouping,
QuantumLayer,
)
from merlin.datasets import iris
p ## 2. Load and Prepare the Iris Dataset
We’ll use the classic Iris dataset for multi-class classification. This dataset is ideal for comparing different architectures as it provides a simple but non-trivial classification task.
The dataset contains:
4 features: sepal length, sepal width, petal length, petal width
3 classes: Setosa, Versicolor, Virginica
150 samples total (split into train/test sets)
[3]:
train_features, train_labels, train_metadata = iris.get_data_train()
test_features, test_labels, test_metadata = iris.get_data_test()
# Convert data to PyTorch tensors
X_train = torch.FloatTensor(train_features)
y_train = torch.LongTensor(train_labels)
X_test = torch.FloatTensor(test_features)
y_test = torch.LongTensor(test_labels)
print(f"Training samples: {X_train.shape[0]}")
print(f"Test samples: {X_test.shape[0]}")
print(f"Features: {X_train.shape[1]}")
print(f"Classes: {len(torch.unique(y_train))}")
Training samples: 120
Test samples: 30
Features: 4
Classes: 3
## 3. Define Model Variants
We’ll test several model architectures to compare classical and quantum approaches:
Classical Models:
MLP: Multi-layer perceptron with 8 hidden units, ReLU activation, and 0.1 dropout
Quantum Models:
LINEAR-7modes-nobunching: Uses linear output mapping with 7 quantum modes and no-bunching constraintLEXGROUPING-7modes: Uses lexicographic grouping strategy for output mappingMODGROUPING-7modes: Uses modular grouping strategy for output mapping
Each model type has a distinct color and line style for visualization.
[4]:
from dataclasses import dataclass
from typing import Literal
import torch
@dataclass
class MLPConfig:
"""Configuration for Multi-Layer Perceptron"""
hidden_sizes: list[int] = None
dropout: float = 0.0
activation: Literal[
"relu", "tanh", "sigmoid", "leaky_relu", "elu", "gelu", "selu"
] = "relu"
normalization: Literal["batch", "layer"] | None = None
class MLP(nn.Module):
"""
An enhanced Multi-Layer Perceptron (MLP) implementation with customizable options.
Features:
- Multiple activation functions
- Batch/Layer normalization options
- Customizable weight initialization
Args:
input_size (int): Dimension of the input features
output_size (int): Dimension of the output
config (MLPConfig): Configuration for the network architecture
"""
# Class-level mapping of available activation functions
ACTIVATIONS = {
"relu": nn.ReLU(),
"tanh": nn.Tanh(),
"sigmoid": nn.Sigmoid(),
"leaky_relu": nn.LeakyReLU(),
"elu": nn.ELU(),
"gelu": nn.GELU(),
"selu": nn.SELU(),
}
def __init__(self, input_size: int, output_size: int, config: MLPConfig):
super().__init__()
last_layer_size = input_size
layers = []
self.config = config
# Validate activation function
if config.activation not in self.ACTIVATIONS:
raise ValueError(f"Unsupported activation function: {config.activation}")
# Build network architecture
for layer_size in config.hidden_sizes:
# Add linear layer
layers.append(nn.Linear(last_layer_size, layer_size))
# Add normalization if specified
if config.normalization == "batch":
layers.append(nn.BatchNorm1d(layer_size))
elif config.normalization == "layer":
layers.append(nn.LayerNorm(layer_size))
# Add activation function
layers.append(self.ACTIVATIONS[config.activation])
# Add dropout if specified
if config.dropout > 0:
layers.append(nn.Dropout(config.dropout))
last_layer_size = layer_size
# Add final output layer
layers.append(nn.Linear(last_layer_size, output_size))
# Create sequential model from layers
self.network = nn.Sequential(*layers)
def initialize_weights(self, method="xavier"):
"""
Initialize network weights using the specified method.
Args:
method (str): Initialization method ('xavier' or 'kaiming') (default: 'xavier')
"""
for layer in self.network:
if isinstance(layer, nn.Linear):
if method == "xavier":
nn.init.xavier_uniform_(layer.weight)
nn.init.zeros_(layer.bias)
elif method == "kaiming":
nn.init.kaiming_normal_(layer.weight, nonlinearity="relu")
nn.init.zeros_(layer.bias)
else:
raise ValueError(f"Unsupported initialization method: {method}")
def forward(self, x):
"""
Forward pass of the network.
Args:
x (torch.Tensor): Input tensor with shape (batch_size, input_size)
Returns:
torch.Tensor: Output tensor with shape (batch_size, output_size)
"""
return self.network(x)
[5]:
def get_model_variants():
"""Define different variants for each model type"""
# Define consistent colors for each model type
MODEL_COLORS = {
"MLP": "#1f77b4", # Blue
"LINEAR": "#2ca02c", # Green
"GROUPING": "#ff7f0e", # Orange
}
# Define line styles for variants
LINE_STYLES = ["--", "-", ":", "-."]
variants = {
"MLP": [
{
"name": "MLP",
"config": MLPConfig(hidden_sizes=[8], dropout=0.1, activation="relu"),
"color": MODEL_COLORS["MLP"],
"linestyle": LINE_STYLES[0],
}
],
"LINEAR": [
{
"name": "LINEAR-7modes-nobunching",
"config": {
"m": 7,
"post_processing": "linear",
"no_bunching": True,
},
"color": MODEL_COLORS["LINEAR"],
"linestyle": LINE_STYLES[1],
}
],
"GROUPING": [
{
"name": "LEXGROUPING-7modes",
"config": {
"m": 6,
"post_processing": "lex",
},
"color": MODEL_COLORS["GROUPING"],
"linestyle": LINE_STYLES[2],
},
{
"name": "MODGROUPING-7modes",
"config": {
"m": 6,
"post_processing": "mod",
},
"color": MODEL_COLORS["GROUPING"],
"linestyle": LINE_STYLES[1],
},
],
}
return variants
## 4. Quantum Circuit Architecture
The quantum circuit is composed of three main sections:
Left interferometer (WL): Performs initial quantum state transformation using beam splitters and phase shifters
Variable phase shifters: Encodes the 4 input features into quantum phases
Right interferometer (WR): Performs final quantum state transformation
The interferometers use a rectangular arrangement of optical elements, where each layer consists of beam splitters (BS) and programmable phase shifters (PS). The phase shifters contain trainable parameters that are optimized during training.
[6]:
def create_quantum_circuit(m):
"""Create quantum circuit with specified number of modes
Parameters:
-----------
m : int
Number of quantum modes in the circuit
Returns:
--------
pcvl.Circuit
Complete quantum circuit with trainable parameters
"""
# Left interferometer with trainable parameters
wl = pcvl.GenericInterferometer(
m,
lambda i: (
pcvl.BS()
// pcvl.PS(pcvl.P(f"theta_li{i}"))
// pcvl.BS()
// pcvl.PS(pcvl.P(f"theta_lo{i}"))
),
shape=pcvl.InterferometerShape.RECTANGLE,
)
# Variable phase shifters for input encoding
c_var = pcvl.Circuit(m)
for i in range(4): # 4 input features
px = pcvl.P(f"px{i + 1}")
c_var.add(i + (m - 4) // 2, pcvl.PS(px))
# Right interferometer with trainable parameters
wr = pcvl.GenericInterferometer(
m,
lambda i: (
pcvl.BS()
// pcvl.PS(pcvl.P(f"theta_ri{i}"))
// pcvl.BS()
// pcvl.PS(pcvl.P(f"theta_ro{i}"))
),
shape=pcvl.InterferometerShape.RECTANGLE,
)
# Combine all components
c = pcvl.Circuit(m)
c.add(0, wl, merge=True)
c.add(0, c_var, merge=True)
c.add(0, wr, merge=True)
return c
## 5. Model Factory Functions
These utility functions handle model creation and parameter counting. The create_model function serves as a factory that instantiates either classical MLP models or quantum models based on the specified configuration.
[7]:
def create_model(model_type, variant):
"""Create model instance based on type and variant
Parameters:
-----------
model_type : str
Type of model ('MLP' or quantum model types)
variant : dict
Configuration dictionary for the model variant
Returns:
--------
nn.Module
PyTorch model instance
"""
if model_type == "MLP":
return MLP(input_size=4, output_size=3, config=variant["config"])
else:
m = variant["config"]["m"]
no_bunching = variant["config"].get("no_bunching", False)
post_processing = variant["config"].get("post_processing")
c = create_quantum_circuit(m)
quantum_layer = QuantumLayer(
input_size=4,
circuit=c,
trainable_parameters=["theta"],
input_parameters=["px"],
input_state=[1, 0] * (m // 2) + [0] * (m % 2),
measurement_strategy=MeasurementStrategy.probs(
computation_space=ComputationSpace.UNBUNCHED
if no_bunching
else ComputationSpace.FOCK
),
)
if post_processing == "linear":
head = nn.Linear(quantum_layer.output_size, 3)
return nn.Sequential(quantum_layer, head)
if post_processing == "lex":
return nn.Sequential(
quantum_layer,
LexGrouping(quantum_layer.output_size, 3),
)
if post_processing == "mod":
return nn.Sequential(
quantum_layer,
ModGrouping(quantum_layer.output_size, 3),
)
return quantum_layer
def count_parameters(model):
"""Count trainable parameters in a PyTorch model"""
return sum(p.numel() for p in model.parameters() if p.requires_grad)
## 6. Training Function
The training function implements a standard supervised learning loop with the following characteristics:
Optimizer: Adam with learning rate 0.02
Loss function: Cross-entropy for multi-class classification
Batch size: 32 samples
Epochs: 50 (default)
The function tracks training loss, training accuracy, and test accuracy at each epoch. After training completes, it generates a detailed classification report.
[8]:
def train_model(
model,
X_train,
y_train,
X_test,
y_test,
model_name,
n_epochs=50,
batch_size=32,
lr=0.02,
):
"""Train a model and track metrics
Parameters:
-----------
model : nn.Module
Model to train
X_train, y_train : torch.Tensor
Training data and labels
X_test, y_test : torch.Tensor
Test data and labels
model_name : str
Name for progress bar display
n_epochs : int
Number of training epochs
batch_size : int
Batch size for mini-batch training
lr : float
Learning rate
Returns:
--------
dict
Training results including losses, accuracies, and final report
"""
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
criterion = nn.CrossEntropyLoss()
losses = []
train_accuracies = []
test_accuracies = []
model.train()
pbar = tqdm(range(n_epochs), leave=False, desc=f"Training {model_name}")
for _epoch in pbar:
# Shuffle training data
permutation = torch.randperm(X_train.size()[0])
total_loss = 0
# Mini-batch training
for i in range(0, X_train.size()[0], batch_size):
indices = permutation[i : i + batch_size]
batch_x, batch_y = X_train[indices], y_train[indices]
outputs = model(batch_x)
loss = criterion(outputs, batch_y)
optimizer.zero_grad()
loss.backward()
optimizer.step()
total_loss += loss.item()
avg_loss = total_loss / (X_train.size()[0] // batch_size)
losses.append(avg_loss)
pbar.set_description(f"Training {model_name} - Loss: {avg_loss:.4f}")
# Evaluation
model.eval()
with torch.no_grad():
# Training accuracy
train_outputs = model(X_train)
train_preds = torch.argmax(train_outputs, dim=1).numpy()
train_acc = accuracy_score(y_train.numpy(), train_preds)
train_accuracies.append(train_acc)
# Test accuracy
test_outputs = model(X_test)
test_preds = torch.argmax(test_outputs, dim=1).numpy()
test_acc = accuracy_score(y_test.numpy(), test_preds)
test_accuracies.append(test_acc)
model.train()
# Generate final classification report
model.eval()
with torch.no_grad():
final_test_outputs = model(X_test)
final_test_preds = torch.argmax(final_test_outputs, dim=1).numpy()
final_report = classification_report(y_test.numpy(), final_test_preds)
return {
"losses": losses,
"train_accuracies": train_accuracies,
"test_accuracies": test_accuracies,
"final_test_acc": test_accuracies[-1],
"classification_report": final_report,
}
## 7. Multi-Run Training Function
To ensure robust and statistically meaningful results, we train each model variant multiple times with different random initializations. This approach helps us:
Understand the variance in model performance
Identify which architectures are more stable
Avoid drawing conclusions from lucky/unlucky single runs
Calculate confidence intervals for performance metrics
The function tracks both individual run results and aggregate statistics across all runs.
[9]:
def train_all_variants(X_train, y_train, X_test, y_test, num_runs=5):
"""Train all model variants multiple times and return results
Parameters:
-----------
X_train, y_train : torch.Tensor
Training data and labels
X_test, y_test : torch.Tensor
Test data and labels
num_runs : int
Number of independent training runs per variant
Returns:
--------
tuple
(all_results, best_models) - Complete results and best performing models
"""
variants = get_model_variants()
all_results = defaultdict(dict)
best_models = {}
for model_type, model_variants in variants.items():
print(f"\n\nTraining {model_type} variants:")
best_acc = 0
for variant in model_variants:
print(
f"\nTraining {variant['name']}... ({count_parameters(create_model(model_type, variant))} parameters)"
)
# Store results from multiple runs
variant_runs = []
for run in range(num_runs):
model = create_model(model_type, variant)
print(f" Run {run + 1}/{num_runs}...")
results = train_model(
model,
X_train,
y_train,
X_test,
y_test,
f"{variant['name']}-run{run + 1}",
)
results["model"] = model
results["color"] = variant["color"]
results["linestyle"] = variant["linestyle"]
variant_runs.append(results)
# Track best model for each type
if results["final_test_acc"] > best_acc:
best_acc = results["final_test_acc"]
best_models[model_type] = {
"name": variant["name"],
"model": model,
"results": results,
}
# Store all runs for this variant
all_results[model_type][variant["name"]] = {
"runs": variant_runs,
"color": variant["color"],
"linestyle": variant["linestyle"],
"avg_final_test_acc": sum(run["final_test_acc"] for run in variant_runs)
/ num_runs,
}
return all_results, best_models
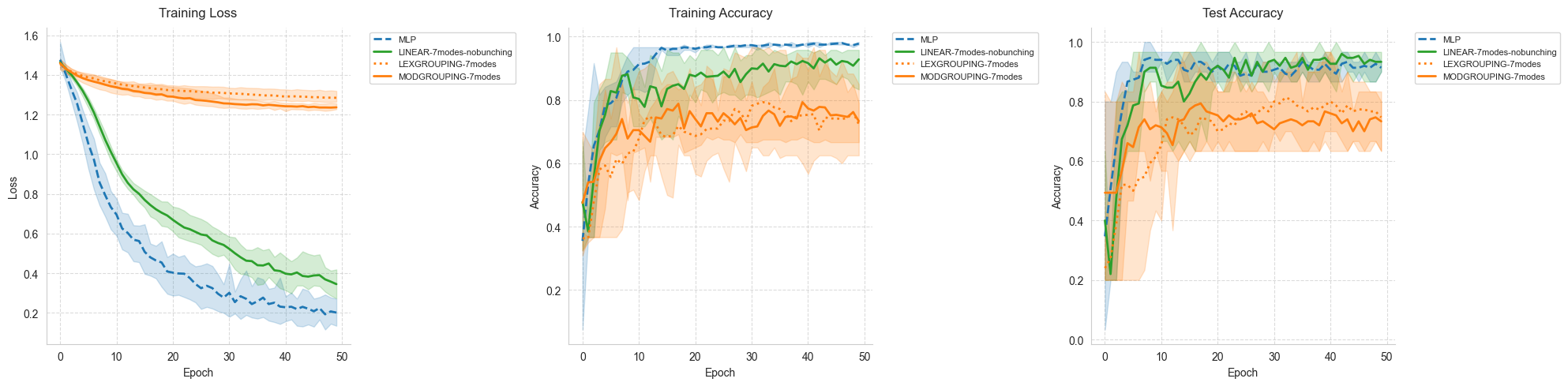
## 8. Visualization Functions
### Training Curves Visualization
This function creates a comprehensive view of the training process by plotting three key metrics:
Training Loss: Shows how well the model fits the training data
Training Accuracy: Indicates the model’s performance on training samples
Test Accuracy: Reveals the model’s generalization capability
For each model variant, we display:
Solid line: Mean performance across all runs
Shaded area: Performance envelope (min to max values)
This visualization helps identify overfitting, convergence speed, and stability.
[10]:
def plot_training_curves(all_results):
"""Plot training curves for all model variants with average and envelope
Shows three subplots:
1. Training loss over epochs
2. Training accuracy over epochs
3. Test accuracy over epochs
Each line represents the mean across runs, with shaded areas showing min/max envelope.
"""
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(20, 5))
# Plot each metric
for _model_type, variants in all_results.items():
for variant_name, variant_data in variants.items():
color = variant_data["color"]
linestyle = variant_data["linestyle"]
label = f"{variant_name}"
# Get data from all runs
losses_runs = [run["losses"] for run in variant_data["runs"]]
train_acc_runs = [run["train_accuracies"] for run in variant_data["runs"]]
test_acc_runs = [run["test_accuracies"] for run in variant_data["runs"]]
# Calculate mean values across all runs
epochs = len(losses_runs[0])
mean_losses = [
sum(run[i] for run in losses_runs) / len(losses_runs)
for i in range(epochs)
]
mean_train_acc = [
sum(run[i] for run in train_acc_runs) / len(train_acc_runs)
for i in range(epochs)
]
mean_test_acc = [
sum(run[i] for run in test_acc_runs) / len(test_acc_runs)
for i in range(epochs)
]
# Calculate min and max values for the envelope
min_losses = [min(run[i] for run in losses_runs) for i in range(epochs)]
max_losses = [max(run[i] for run in losses_runs) for i in range(epochs)]
min_train_acc = [
min(run[i] for run in train_acc_runs) for i in range(epochs)
]
max_train_acc = [
max(run[i] for run in train_acc_runs) for i in range(epochs)
]
min_test_acc = [min(run[i] for run in test_acc_runs) for i in range(epochs)]
max_test_acc = [max(run[i] for run in test_acc_runs) for i in range(epochs)]
# Plot mean lines
ax1.plot(
mean_losses, label=label, color=color, linestyle=linestyle, linewidth=2
)
ax2.plot(
mean_train_acc,
label=label,
color=color,
linestyle=linestyle,
linewidth=2,
)
ax3.plot(
mean_test_acc,
label=label,
color=color,
linestyle=linestyle,
linewidth=2,
)
# Plot envelopes (filled area between min and max)
epochs_range = range(epochs)
ax1.fill_between(
epochs_range, min_losses, max_losses, color=color, alpha=0.2
)
ax2.fill_between(
epochs_range, min_train_acc, max_train_acc, color=color, alpha=0.2
)
ax3.fill_between(
epochs_range, min_test_acc, max_test_acc, color=color, alpha=0.2
)
# Customize plots
for ax, title in zip(
[ax1, ax2, ax3],
["Training Loss", "Training Accuracy", "Test Accuracy"],
strict=False,
):
ax.set_title(title, fontsize=12, pad=10)
ax.set_xlabel("Epoch", fontsize=10)
ax.set_ylabel(title.split()[-1], fontsize=10)
ax.legend(fontsize=8, bbox_to_anchor=(1.05, 1), loc="upper left")
ax.grid(True, linestyle="--", alpha=0.7)
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
plt.tight_layout()
plt.show()
### Confusion Matrix Visualization
Confusion matrices provide detailed insights into classification performance by showing:
Which classes are correctly classified (diagonal elements)
Which classes are confused with each other (off-diagonal elements)
Overall performance patterns for each model architecture
We display the confusion matrix for the best-performing model of each type.
[11]:
def plot_best_confusion_matrices(best_models, X_test, y_test):
"""Plot confusion matrices for the best model of each type"""
fig, axes = plt.subplots(1, 3, figsize=(20, 5))
class_names = ["setosa", "versicolor", "virginica"]
for idx, (model_type, best) in enumerate(best_models.items()):
model = best["model"]
model.eval()
with torch.no_grad():
outputs = model(X_test)
predictions = torch.argmax(outputs, dim=1).numpy()
cm = confusion_matrix(y_test.numpy(), predictions)
sns.heatmap(
cm,
annot=True,
fmt="d",
cmap="Blues",
xticklabels=class_names,
yticklabels=class_names,
ax=axes[idx],
)
axes[idx].set_title(f"Best {model_type}\n({best['name']})")
axes[idx].set_xlabel("Predicted")
axes[idx].set_ylabel("True")
plt.setp(axes[idx].get_xticklabels(), rotation=45)
plt.setp(axes[idx].get_yticklabels(), rotation=45)
plt.tight_layout()
plt.show()
## 9. Results Summary Function
This function provides a comprehensive statistical analysis of the experimental results, including:
Parameter efficiency: Number of trainable parameters for each model
Performance statistics: Mean accuracy, standard deviation, min/max values
Detailed classification reports: Precision, recall, and F1-score for each class
These metrics help in making informed decisions about model selection based on the specific requirements of your application.
[12]:
def print_comparison_results(all_results, best_models):
"""Print detailed comparison of all models and variants with multi-run statistics"""
print("\n----- Model Comparison Results -----")
# Print results for all variants
print("\nAll Model Variants Results (averaged over multiple runs):")
for model_type, variants in all_results.items():
print(f"\n{model_type} Variants:")
for variant_name, variant_data in variants.items():
print(f"\n{variant_name}:")
print(f"Parameters: {count_parameters(variant_data['runs'][0]['model'])}")
# Calculate statistics across runs
final_accs = [run["final_test_acc"] for run in variant_data["runs"]]
avg_acc = sum(final_accs) / len(final_accs)
min_acc = min(final_accs)
max_acc = max(final_accs)
std_acc = (
sum((acc - avg_acc) ** 2 for acc in final_accs) / len(final_accs)
) ** 0.5
print(
f"Final Test Accuracy: {avg_acc:.4f} ± {std_acc:.4f} (min: {min_acc:.4f}, max: {max_acc:.4f})"
)
# Print best model results
print("\nBest Models:")
for model_type, best in best_models.items():
print(f"\nBest {model_type} Model: {best['name']}")
print(f"Final Test Accuracy: {best['results']['final_test_acc']:.4f}")
print(f"Classification Report:\n{best['results']['classification_report']}")
## 10. Run the Complete Experiment
Now we’ll execute the full experimental pipeline. The experiment consists of:
Multiple training runs: Each model variant is trained 5 times with different random seeds
Performance tracking: All metrics are recorded for statistical analysis
Visualization: Training curves and confusion matrices are generated
Statistical summary: Detailed performance statistics are computed
This comprehensive approach ensures reliable and reproducible results.
[13]:
# Set number of independent runs per model variant
num_runs = 5
# Train all variants
print("Starting training process...")
all_results, best_models = train_all_variants(
X_train, y_train, X_test, y_test, num_runs=num_runs
)
Starting training process...
Training MLP variants:
Training MLP... (67 parameters)
Run 1/5...
Run 2/5...
Run 3/5...
Run 4/5...
Run 5/5...
Training LINEAR variants:
Training LINEAR-7modes-nobunching... (192 parameters)
Run 1/5...
Run 2/5...
Run 3/5...
Run 4/5...
Run 5/5...
Training GROUPING variants:
Training LEXGROUPING-7modes... (60 parameters)
Run 1/5...
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
Run 2/5...
Run 3/5...
Run 4/5...
Run 5/5...
Training MODGROUPING-7modes... (60 parameters)
Run 1/5...
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
Run 2/5...
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
Run 3/5...
Run 4/5...
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
Run 5/5...
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
/Users/lfvigneux/miniconda3/envs/MerLin_dev/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1833: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", result.shape[0])
### Visualize Training Progress
The following plots show how each model’s performance evolves during training. Pay attention to:
Convergence speed: How quickly each model reaches its best performance
Stability: Width of the shaded area indicates variance across runs
Final performance: Where each model plateaus
[14]:
plot_training_curves(all_results)
### Confusion Matrices for Best Models
These confusion matrices reveal the classification patterns of the best-performing model from each architecture type. Look for:
Perfect classification: Bright blue diagonal with zeros elsewhere
Common confusions: Off-diagonal elements show which classes are hard to distinguish
[15]:
print_comparison_results(all_results, best_models)
----- Model Comparison Results -----
All Model Variants Results (averaged over multiple runs):
MLP Variants:
MLP:
Parameters: 67
Final Test Accuracy: 0.9267 ± 0.0249 (min: 0.9000, max: 0.9667)
LINEAR Variants:
LINEAR-7modes-nobunching:
Parameters: 192
Final Test Accuracy: 0.9200 ± 0.0452 (min: 0.8667, max: 0.9667)
GROUPING Variants:
LEXGROUPING-7modes:
Parameters: 60
Final Test Accuracy: 0.8333 ± 0.1116 (min: 0.6333, max: 0.9667)
MODGROUPING-7modes:
Parameters: 60
Final Test Accuracy: 0.6733 ± 0.0800 (min: 0.6333, max: 0.8333)
Best Models:
Best MLP Model: MLP
Final Test Accuracy: 0.9667
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 13
1 0.86 1.00 0.92 6
2 1.00 0.91 0.95 11
accuracy 0.97 30
macro avg 0.95 0.97 0.96 30
weighted avg 0.97 0.97 0.97 30
Best LINEAR Model: LINEAR-7modes-nobunching
Final Test Accuracy: 0.9667
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 13
1 0.86 1.00 0.92 6
2 1.00 0.91 0.95 11
accuracy 0.97 30
macro avg 0.95 0.97 0.96 30
weighted avg 0.97 0.97 0.97 30
Best GROUPING Model: LEXGROUPING-7modes
Final Test Accuracy: 0.9667
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 13
1 0.86 1.00 0.92 6
2 1.00 0.91 0.95 11
accuracy 0.97 30
macro avg 0.95 0.97 0.96 30
weighted avg 0.97 0.97 0.97 30
## 11. Key Findings and Conclusions
Based on the experimental results, we can draw several important conclusions:
Model Complexity vs Performance:
Compare the number of parameters required by each architecture
Assess whether quantum models achieve similar performance with fewer parameters
Training Stability:
The variance across multiple runs indicates how sensitive each model is to initialization
Lower variance suggests more reliable training
Generalization Capability:
The gap between training and test accuracy reveals overfitting tendencies
Smaller gaps indicate better generalization
Classification Patterns:
Confusion matrices show which flower species are most difficult to distinguish
This can guide feature engineering or model selection
Practical Considerations:
For deployment, consider:
Classical MLPs: Well-understood, fast inference, established deployment pipelines
Quantum models: Potentially more parameter-efficient, may offer advantages on quantum hardware
Future Research Directions:
Scaling to larger datasets: Test on more complex classification tasks
Noise modeling: Investigate performance under realistic quantum noise conditions
Hybrid architectures: Combine classical and quantum layers
Hardware implementation: Evaluate on actual quantum photonic devices
Feature encoding strategies: Explore different ways to encode classical data into quantum states